Conformational dynamics of membrane transporters
Transporters are essential molecules responsible for the movement of substrates across biological membranes. Our understanding of the range of conformations adopted by transporters in order to carry out their essential biologica functions remains limited as it can be very difficult to obtain information on these using conventional structural biology approaches. The challenge is even greater for eukaryotic systems due to their intrinsic dynamics and the often low yield of purification. The group employs the emerging Hydrogen Deuterium Exchange-Mass Spectrometry (HDX-MS) technology to capture the conformational states of transporters including, sugar transporters (XylE, LacY) and the eukaryotic transporter, UapA, within its native lipid composition.
We have established mechanistic details on how the lipid environment affects the conformational dynamics of sugar transporters. This has also led to building a comprehensive HDX-MS protocol for carrying out experiments in lipid nanodiscs and combining them molecular dynamics simulations. We have showed how ligand binding impacts structural dynamics and also establish the role of lipids on oligomeric states of eukaryotic transporters.

Links to publications: 1, 2, 3, 4, 5
Probing the conformational landscape of GPCRs
Biological membranes form an essential barrier between living cells and their external environments. Membrane proteins (MPs) are the gateways within these structures with essential functions in signalling and transport. While powerful structural techniques such as cryo-EM and X-ray crystallography have offered invaluable information on MPs structure and function in recent years, these techniques are limited to static snapshots of isolated conformational states. They also tend not to account for surrounding lipid environments precluding our capacity to characterize dynamics in native environments. This is particularly pertinent for eukaryotic proteins that are embedded in heterogenous lipid environments and are often bottle-necked by low purification yields making them impractical to study by structural methods.
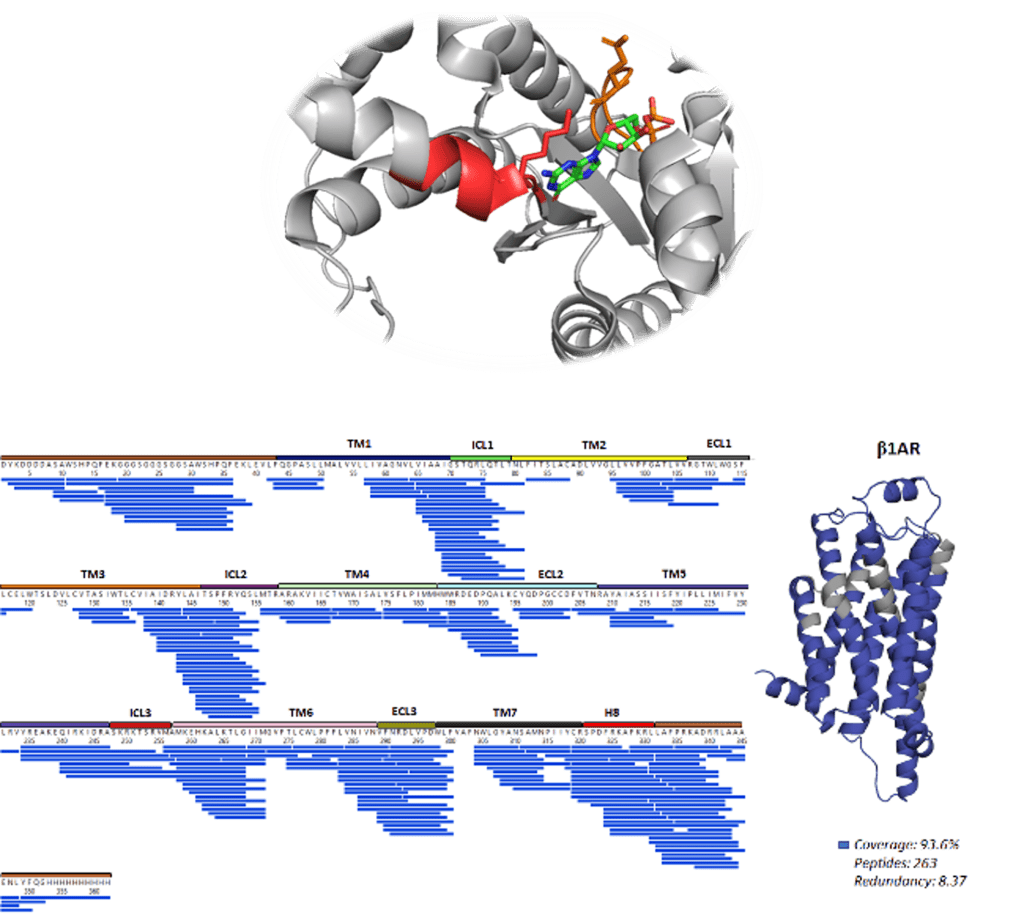
Hydrogen deuterium exchange mass spectrometry (HDX-MS) offers a highly sensitive tool to interrogate the structural dynamics of MPs by monitoring the exchange of hydrogen to bulk deuterium at native temperature. More recently, our group and others have developed approaches to monitor structural changes of eukaryotic proteins including G protein-coupled receptors (GPCRs). HDX-MS has been employed to characterise the structural dynamics of turkey β1-adrenergic receptor (tβ1AR) in complex with nine ligands, including agonists, partial agonists and antagonists. We show that dynamic signatures across the GPCR structure can be grouped by compound modality. Surprisingly, we discovered repeated destabilisation of the intracellular loop 1 (ICL1) upon full agonist binding and stabilisation upon antagonist binding, suggesting that increased dynamics in this region are an essential component for G-protein recruitment.
We also target the neurotensin receptor NTSR1, a GPCR responsible for mediating responses to neurotensin (NTS) and is implicated in a wide range of physiological processes including blood pressure, ileum contraction or relaxation, analgesia and hypothermia. This approach is now to be expanded to study of NTSR1 in lipidic nanodiscs to characterise protein dynamics with endogenous lipids and ligands.
Structure and dynamics of the bacterial secretesome (collaboration with Ian Collinson, University of Bristol)
The general secretory machinery is essential for every cell of every organism. The hetero-trimeric core-complex (SecYEG in bacteria) forms the protein-channel across and into phospholipid bilayers, respectively for protein secretion and membrane insertion. Many factors associate with this core-translocon to drive and regulate translocation, and for the reasons of quality control. Bacteria, for example, deploy SecA for ATP- and proton-motive-force (PMF)-driven post-translational transport across the inner-membrane, while insertion is predominantly delivered by associated translating ribosomes. SecYEG also associates with ancillary factors forming the holo-translocon (HTL) wherein the SecDF sub-complex promotes secretion) and YidC facilitates trans-membrane helix (TMH) insertion.
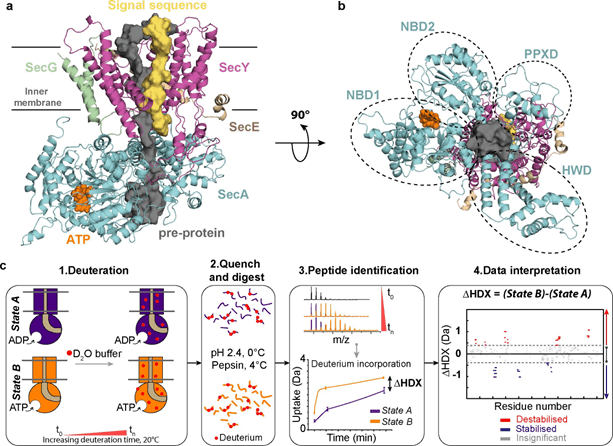
HDX-MS has been used to study the dynamic behaviour of the components of the secretosome, which has already been tried and tested with sub-components of the secretory machinery. HDX-MS constitutes a very sensitive tool that reports on protein dynamics by tracking the exchange of backbone hydrogen atoms for deuterium at peptide-level resolution. Recent, pioneering workflows introduced by the Politis lab and others have enabled analysis of membrane proteins and other complex biomolecules in native-like conditions. Particularly, HDX-MS offers advantages over traditional approaches; it tolerates heterogeneous environments (e.g. membranes, vesicles), has low sample requirements (from high nM to low μM range) with no need for chemical probes or bio-orthogonal labels. Importantly, HDX-MS data report upon the conformational equilibrium of protein ensembles including all relevant populations. As such, HDX-MS has emerged as a powerful tool to study dynamic mechanisms often inaccessible by other structural techniques. This emerging technique is the beneficiary of rapid and unrelenting technological advances in instrumentation, such as the state-of-the-art cyclic ion mobility HDX mass spectrometer that are deployed in HDX-MS experiments.
Links to publications: 1, 2, 3
